The majority of figures presented here are found in published journal articles, referenced here. The publications page lists additional articles for projects I supported.
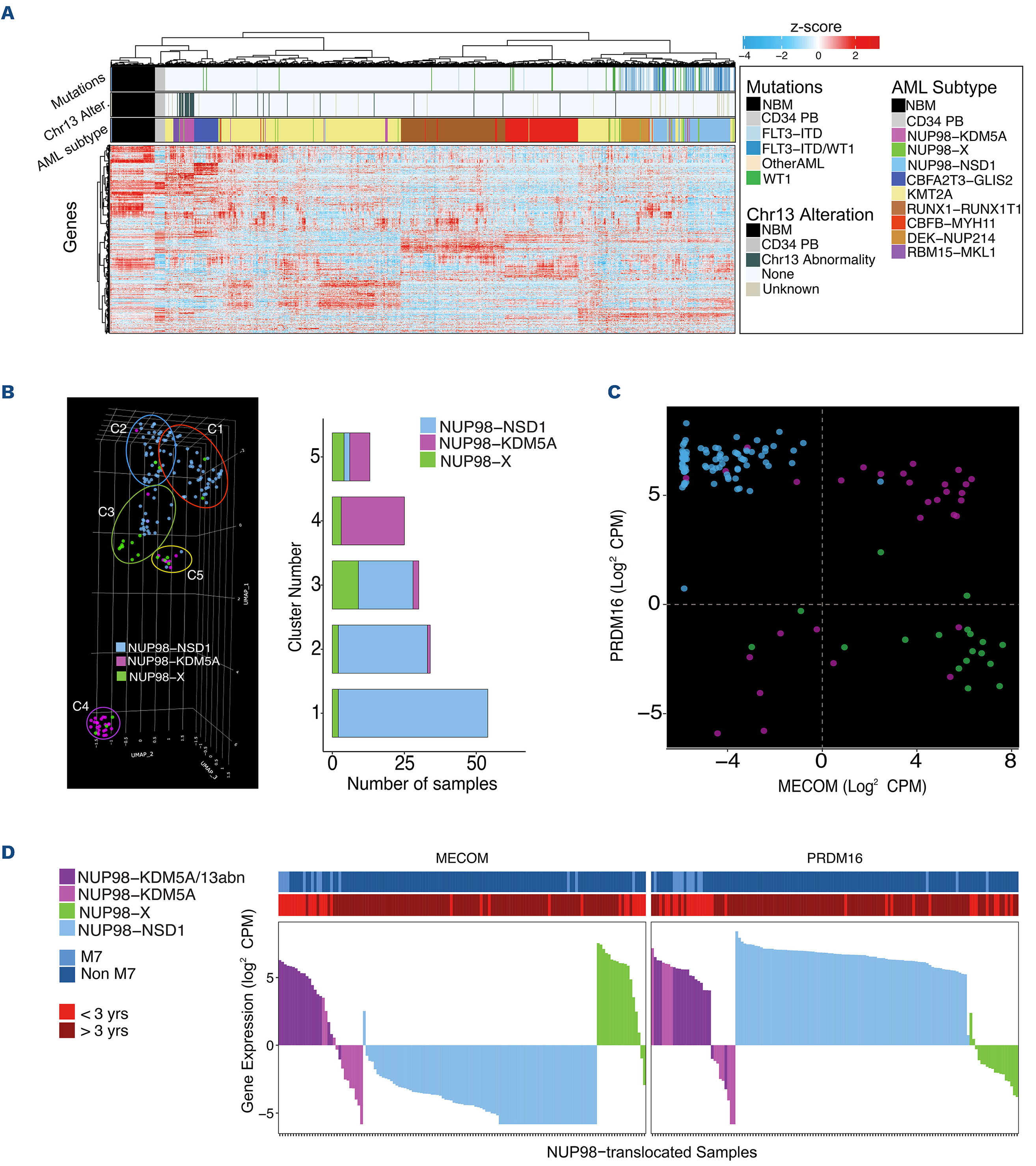
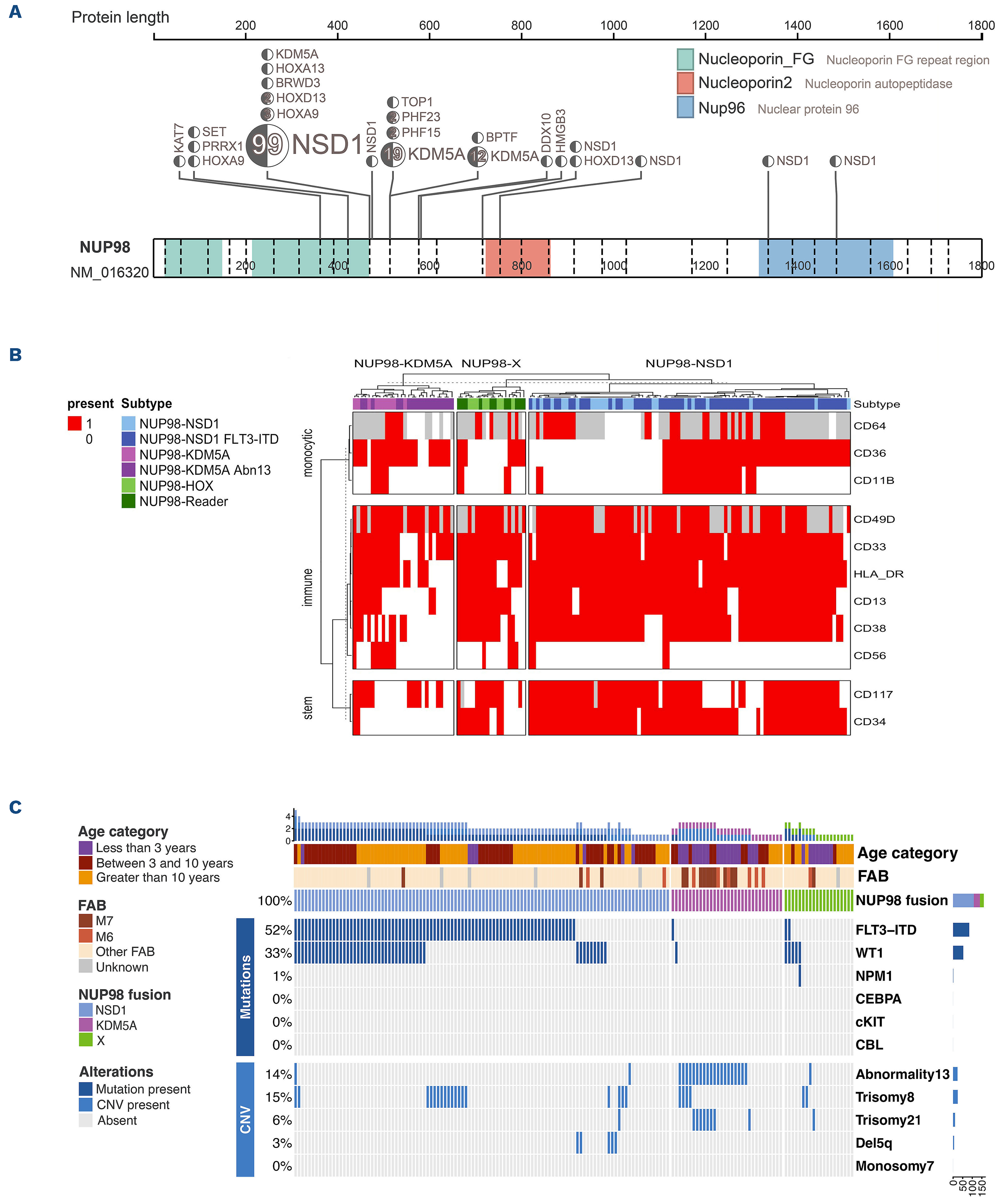
Bertrums, E. J. M., Smith, J. L., Harmon, L., Ries, R. E., Wang, Y.-C. J., Alonzo, T. A., Menssen, A. J., Chisholm, K. M., Leonti, A. R., Tarlock, K., Ostronoff, F., Pogosova-Agadjanyan, E. L., Kaspers, G. J. L., Hasle, H., Dworzak, M., Walter, C., Muhlegger, N., Morerio, C., Pardo, L., … Meshinchi, S. (2023). Comprehensive molecular and clinical characterization of
NUP98 fusions in pediatric acute myeloid leukemia.
Haematologica.
https://doi.org/10.3324/haematol.2022.281653
Chisholm, K. M., Smith, J., Heerema-McKenney, A. E., Choi, J. K., Ries, R. E., Hirsch, B. A., Raimondi, S. C., Wang, Y.-C., Dang, A., Alonzo, T. A., Sung, L., Aplenc, R., Gamis, A. S., Meshinchi, S., & Kahwash, S. B. (2023). Pathologic, cytogenetic, and molecular features of acute myeloid leukemia with megakaryocytic differentiation: A report from the children
s oncology group.
Pediatric Blood & Cancer.
https://doi.org/10.1002/pbc.30251
Farrar, J. E., Smith, J. L., Othus, M., Huang, B. J., Wang, Y.-C., Ries, R., Hylkema, T., Pogosova-Agadjanyan, E. L., Challa, S., Leonti, A., Shaw, T. I., Triche, T. J., Gamis, A. S., Aplenc, R., Kolb, E. A., Ma, X., Stirewalt, D. L., Alonzo, T. A., & Meshinchi, S. (2023). Long noncoding RNA expression independently predicts outcome in pediatric acute myeloid leukemia.
Journal of Clinical Oncology,
41(16), 2949–2962.
https://doi.org/10.1200/jco.22.01114
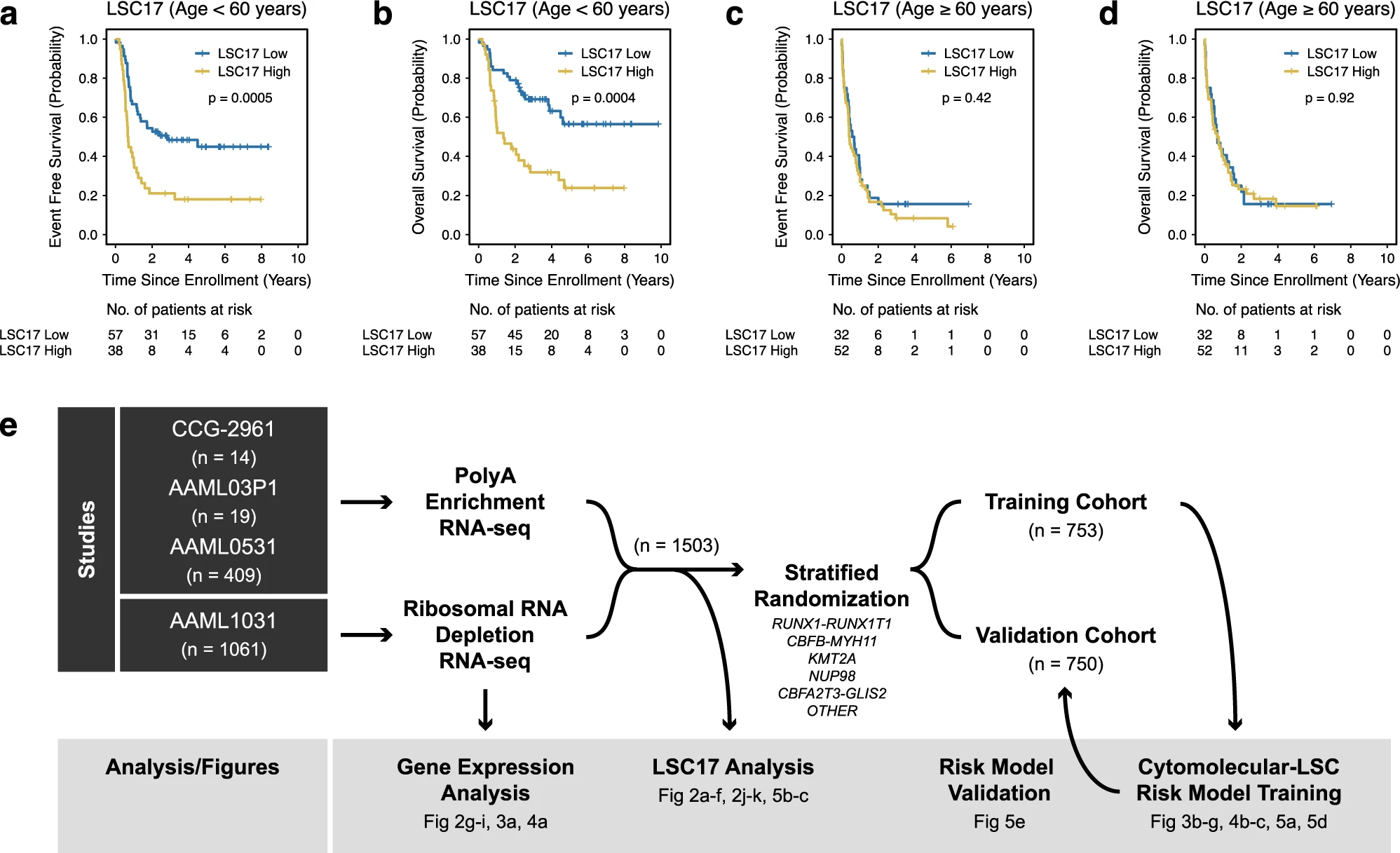
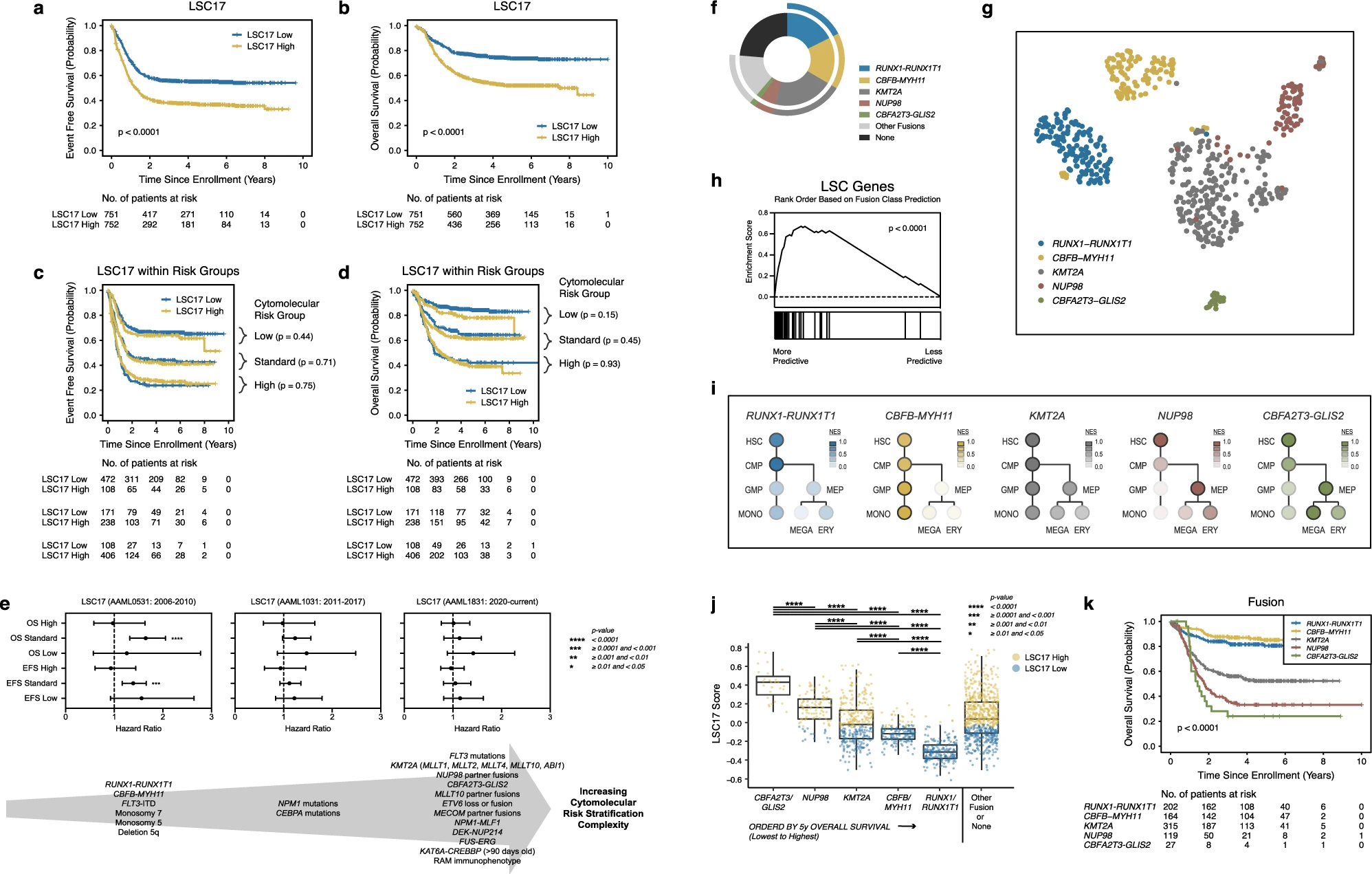
Huang, B. J., Smith, J. L., Farrar, J. E., Wang, Y.-C., Umeda, M., Ries, R. E., Leonti, A. R., Crowgey, E., Furlan, S. N., Tarlock, K., Armendariz, M., Liu, Y., Shaw, T. I., Wei, L., Gerbing, R. B., Cooper, T. M., Gamis, A. S., Aplenc, R., Kolb, E. A., … Meshinchi, S. (2022). Integrated stem cell signature and cytomolecular risk determination in pediatric acute myeloid leukemia.
Nature Communications,
13(1).
https://doi.org/10.1038/s41467-022-33244-6
Huang, B. J., Smith, J. L., Wang, Y.-C., Taghizadeh, K., Leonti, A. R., Ries, R. E., Liu, Y., Kolekar, P., Tarlock, K., Gerbing, R., Crowgey, E., Furlan, S. N., Shaw, T. I., Hagiwara, K., Wei, L., Cooper, T. M., Gamis, A. S., Aplenc, R., Kolb, E. A., … Meshinchi, S. (2021). CBFB-MYH11 fusion transcripts distinguish acute myeloid leukemias with distinct molecular landscapes and outcomes.
Blood Advances,
5(23), 4963–4968.
https://doi.org/10.1182/bloodadvances.2021004965
Kappe, S., Zanghi, G., Patel, H., Camargo, N., Smith, J., Bae, Y., Hesping, E., Boddey, J., Flannery, E., Chuenchob, V., Fishbaugher, M., Mikolajczak, S., Roobsoong, W., Sattabongkot, J., Hayes, K., & Vaughan, A. (2023).
Genome-wide gene expression profiles throughout human malaria parasite liver stage development.
https://doi.org/10.21203/rs.3.rs-3689897/v1
Le, Q., Hadland, B., Smith, J. L., Leonti, A., Huang, B. J., Ries, R., Hylkema, T. A., Castro, S., Tang, T. T., McKay, C. N., Perkins, L., Pardo, L., Sarthy, J., Beckman, A. K., Williams, R., Idemmili, R., Furlan, S., Ishida, T., Call, L., … Meshinchi, S. (2022).
CBFA2T3-
GLIS2 model of pediatric acute megakaryoblastic leukemia identifies
FOLR1 as a
CAR t cell target.
Journal of Clinical Investigation,
132(22).
https://doi.org/10.1172/jci157101
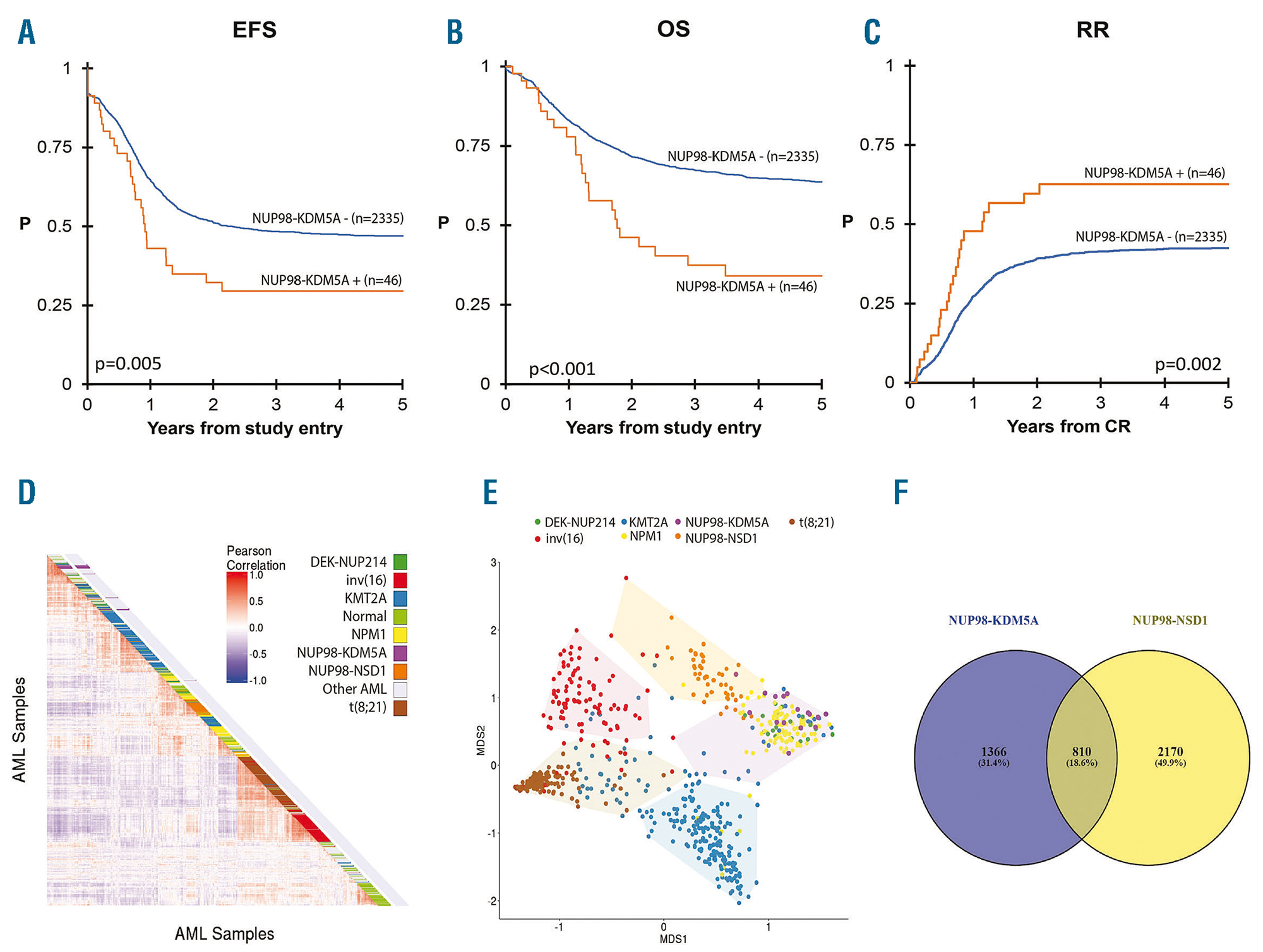
Noort, S., Wander, P., Alonzo, T. A., Smith, J., Ries, R. E., Gerbing, R. B., Dolman, M. E. M., Locatelli, F., Reinhardt, D., Baruchel, A., Stary, J., Molenaar, J. J., Stam, R. W., Heuvel-Eibrink, M. M. van den, Zwaan, C. M., & Meshinchi, S. (2020). The clinical and biological characteristics of
NUP98-
KDM5A in pediatric acute myeloid leukemia.
Haematologica, haematol.2019.236745.
https://doi.org/10.3324/haematol.2019.236745
Noort, S., Zimmermann, M., Reinhardt, D., Cuccuini, W., Pigazzi, M., Smith, J., Ries, R. E., Alonzo, T. A., Hirsch, B., Tomizawa, D., Locatelli, F., Gruber, T. A., Raimondi, S., Sonneveld, E., Cheuk, D. K., Dworzak, M., Stary, J., Abrahamsson, J., Arad-Cohen, N., … Zwaan, C. M. (2018). Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric
AML: A retrospective study by the i-
BFM study group.
Blood,
132(15), 1584–1592.
https://doi.org/10.1182/blood-2018-05-849059
Smith, J. L. (2019). Consensus machine learning for gene target selection in pediatric AML risk. In biorxiv.
Smith, J. L., Ries, R. E., Farrar, J. E., Alonzo, T. A., Gerbing, R. B., Leonti, A. R., Hylkema, T. A., Gamis, A. S., Aplenc, R., Kolb, E. A., Nguyen, C., Meerzaman, D., Triche, T. J., & Meshinchi, S. (2019). Long non-coding RNAs (lncRNAs) are highly associated with disease characteristics and outcome in pediatric acute myeloid leukemia - a COG and tpaml study.
Blood,
134(Supplement_1), 2741–2741.
https://doi.org/10.1182/blood-2019-127318
Smith, J. L., Ries, R. E., Wang, Y.-C., Leonti, A. R., Alonzo, T. A., Gamis, A. S., Aplenc, R., Kolb, A. E., Huang, B. J., Ma, X., Shaw, T. I., & Meshinchi, S. (2021). ETS family transcription factor fusions in childhood AML: Distinct expression networks and clinical implications.
Blood,
138(Supplement 1), 2356–2356.
https://doi.org/10.1182/blood-2021-148894
Tarlock, K., Lamble, A., Wang, J., Gerbing, R. B., Ries, R. E., Loken, M. R., Brodersen, L. E., Pardo, L., Leonti, A. R., Smith, J., Hylkema, T., Woods, W. G., Cooper, T., Kolb, E. A., Gamis, A. S., Aplenc, R., Alonzo, T., & Meshinchi, S. (2021).
CEBPA bZip mutations are associated with favorable prognosis in de novo
AML: A report from the children
s oncology group.
Blood.
https://doi.org/10.1182/blood.2020009652
Bertrums, E. J. M., Smith, J. L., Harmon, L., Ries, R. E., Wang, Y.-C. J., Alonzo, T. A., Menssen, A. J., Chisholm, K. M., Leonti, A. R., Tarlock, K., Ostronoff, F., Pogosova-Agadjanyan, E. L., Kaspers, G. J. L., Hasle, H., Dworzak, M., Walter, C., Muhlegger, N., Morerio, C., Pardo, L., … Meshinchi, S. (2023). Comprehensive molecular and clinical characterization of
NUP98 fusions in pediatric acute myeloid leukemia.
Haematologica.
https://doi.org/10.3324/haematol.2022.281653
Chisholm, K. M., Smith, J., Heerema-McKenney, A. E., Choi, J. K., Ries, R. E., Hirsch, B. A., Raimondi, S. C., Wang, Y.-C., Dang, A., Alonzo, T. A., Sung, L., Aplenc, R., Gamis, A. S., Meshinchi, S., & Kahwash, S. B. (2023). Pathologic, cytogenetic, and molecular features of acute myeloid leukemia with megakaryocytic differentiation: A report from the children
s oncology group.
Pediatric Blood & Cancer.
https://doi.org/10.1002/pbc.30251
Farrar, J. E., Smith, J. L., Othus, M., Huang, B. J., Wang, Y.-C., Ries, R., Hylkema, T., Pogosova-Agadjanyan, E. L., Challa, S., Leonti, A., Shaw, T. I., Triche, T. J., Gamis, A. S., Aplenc, R., Kolb, E. A., Ma, X., Stirewalt, D. L., Alonzo, T. A., & Meshinchi, S. (2023). Long noncoding RNA expression independently predicts outcome in pediatric acute myeloid leukemia.
Journal of Clinical Oncology,
41(16), 2949–2962.
https://doi.org/10.1200/jco.22.01114
Huang, B. J., Smith, J. L., Farrar, J. E., Wang, Y.-C., Umeda, M., Ries, R. E., Leonti, A. R., Crowgey, E., Furlan, S. N., Tarlock, K., Armendariz, M., Liu, Y., Shaw, T. I., Wei, L., Gerbing, R. B., Cooper, T. M., Gamis, A. S., Aplenc, R., Kolb, E. A., … Meshinchi, S. (2022). Integrated stem cell signature and cytomolecular risk determination in pediatric acute myeloid leukemia.
Nature Communications,
13(1).
https://doi.org/10.1038/s41467-022-33244-6
Huang, B. J., Smith, J. L., Wang, Y.-C., Taghizadeh, K., Leonti, A. R., Ries, R. E., Liu, Y., Kolekar, P., Tarlock, K., Gerbing, R., Crowgey, E., Furlan, S. N., Shaw, T. I., Hagiwara, K., Wei, L., Cooper, T. M., Gamis, A. S., Aplenc, R., Kolb, E. A., … Meshinchi, S. (2021). CBFB-MYH11 fusion transcripts distinguish acute myeloid leukemias with distinct molecular landscapes and outcomes.
Blood Advances,
5(23), 4963–4968.
https://doi.org/10.1182/bloodadvances.2021004965
Kappe, S., Zanghi, G., Patel, H., Camargo, N., Smith, J., Bae, Y., Hesping, E., Boddey, J., Flannery, E., Chuenchob, V., Fishbaugher, M., Mikolajczak, S., Roobsoong, W., Sattabongkot, J., Hayes, K., & Vaughan, A. (2023).
Genome-wide gene expression profiles throughout human malaria parasite liver stage development.
https://doi.org/10.21203/rs.3.rs-3689897/v1
Le, Q., Hadland, B., Smith, J. L., Leonti, A., Huang, B. J., Ries, R., Hylkema, T. A., Castro, S., Tang, T. T., McKay, C. N., Perkins, L., Pardo, L., Sarthy, J., Beckman, A. K., Williams, R., Idemmili, R., Furlan, S., Ishida, T., Call, L., … Meshinchi, S. (2022).
CBFA2T3-
GLIS2 model of pediatric acute megakaryoblastic leukemia identifies
FOLR1 as a
CAR t cell target.
Journal of Clinical Investigation,
132(22).
https://doi.org/10.1172/jci157101
Noort, S., Wander, P., Alonzo, T. A., Smith, J., Ries, R. E., Gerbing, R. B., Dolman, M. E. M., Locatelli, F., Reinhardt, D., Baruchel, A., Stary, J., Molenaar, J. J., Stam, R. W., Heuvel-Eibrink, M. M. van den, Zwaan, C. M., & Meshinchi, S. (2020). The clinical and biological characteristics of
NUP98-
KDM5A in pediatric acute myeloid leukemia.
Haematologica, haematol.2019.236745.
https://doi.org/10.3324/haematol.2019.236745
Noort, S., Zimmermann, M., Reinhardt, D., Cuccuini, W., Pigazzi, M., Smith, J., Ries, R. E., Alonzo, T. A., Hirsch, B., Tomizawa, D., Locatelli, F., Gruber, T. A., Raimondi, S., Sonneveld, E., Cheuk, D. K., Dworzak, M., Stary, J., Abrahamsson, J., Arad-Cohen, N., … Zwaan, C. M. (2018). Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric
AML: A retrospective study by the i-
BFM study group.
Blood,
132(15), 1584–1592.
https://doi.org/10.1182/blood-2018-05-849059
Smith, J. L. (2019). Consensus machine learning for gene target selection in pediatric AML risk. In biorxiv.
Smith, J. L., Ries, R. E., Farrar, J. E., Alonzo, T. A., Gerbing, R. B., Leonti, A. R., Hylkema, T. A., Gamis, A. S., Aplenc, R., Kolb, E. A., Nguyen, C., Meerzaman, D., Triche, T. J., & Meshinchi, S. (2019). Long non-coding RNAs (lncRNAs) are highly associated with disease characteristics and outcome in pediatric acute myeloid leukemia - a COG and tpaml study.
Blood,
134(Supplement_1), 2741–2741.
https://doi.org/10.1182/blood-2019-127318
Smith, J. L., Ries, R. E., Wang, Y.-C., Leonti, A. R., Alonzo, T. A., Gamis, A. S., Aplenc, R., Kolb, A. E., Huang, B. J., Ma, X., Shaw, T. I., & Meshinchi, S. (2021). ETS family transcription factor fusions in childhood AML: Distinct expression networks and clinical implications.
Blood,
138(Supplement 1), 2356–2356.
https://doi.org/10.1182/blood-2021-148894
Tarlock, K., Lamble, A., Wang, J., Gerbing, R. B., Ries, R. E., Loken, M. R., Brodersen, L. E., Pardo, L., Leonti, A. R., Smith, J., Hylkema, T., Woods, W. G., Cooper, T., Kolb, E. A., Gamis, A. S., Aplenc, R., Alonzo, T., & Meshinchi, S. (2021).
CEBPA bZip mutations are associated with favorable prognosis in de novo
AML: A report from the children
s oncology group.
Blood.
https://doi.org/10.1182/blood.2020009652